Soft active living matter
Soft active living matter
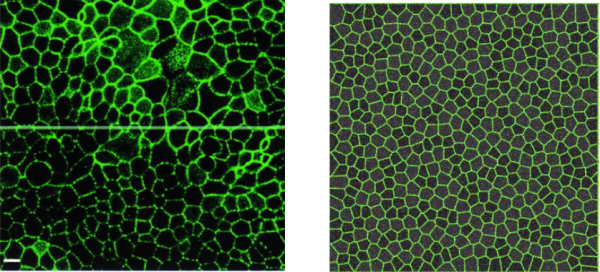 “Vertex” models, which treat dense collections of interacting cells as polygonal or polyhedral decompositions of space, are increasingly prevalent in biophysical modeling of tissues and cellular aggregates. Computational approaches to studying these models must confront a number of challenges; in contrast to standard molecular dynamics simulations of particles interacting via (typically) short-ranged forces, in vertex models the interaction depend on both the geometry and the topology of the interacting units. For instance, in vertex models that treat cells as Voronoi volume elements either a Voronoi or Delaunay triangulation must be maintained at all times in the course of the simulation and forces are calculated based on relative cell position and with reference to the underlying triangulation.
“Vertex” models, which treat dense collections of interacting cells as polygonal or polyhedral decompositions of space, are increasingly prevalent in biophysical modeling of tissues and cellular aggregates. Computational approaches to studying these models must confront a number of challenges; in contrast to standard molecular dynamics simulations of particles interacting via (typically) short-ranged forces, in vertex models the interaction depend on both the geometry and the topology of the interacting units. For instance, in vertex models that treat cells as Voronoi volume elements either a Voronoi or Delaunay triangulation must be maintained at all times in the course of the simulation and forces are calculated based on relative cell position and with reference to the underlying triangulation.
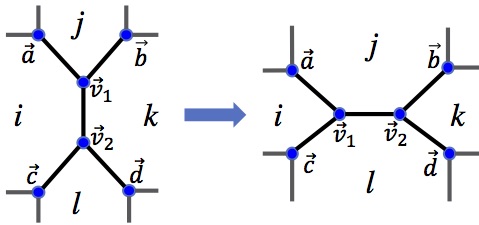 As a result, the most sophisticated implementations of vertex models rely on serial, single-threaded CPU-based implementations. In the absence of the parallelized implementations common in many other simulation frameworks vertex models struggle to tackle both long-timescale problems and simulations of large numbers of interacting cells. I have worked on correcting this deficiency, implementing GPU-accelerated algorithms to study these models, increasing by orders of magnitude the time and length scales accessible to researchers in the field.
As a result, the most sophisticated implementations of vertex models rely on serial, single-threaded CPU-based implementations. In the absence of the parallelized implementations common in many other simulation frameworks vertex models struggle to tackle both long-timescale problems and simulations of large numbers of interacting cells. I have worked on correcting this deficiency, implementing GPU-accelerated algorithms to study these models, increasing by orders of magnitude the time and length scales accessible to researchers in the field.
Related Publications
- cellGPU: GPU-accelerated off-lattice cell simulations
- cellGPU repository: webpage and additional documentation